Identification of novel functional peptides contained in red alga Upppleinori protein
Section outline
-
In our laboratory, we are researching the components contained in red algae as a “search for health functional components in red algae, low-utilization resources”. Among them, peptides derived from proteins of red algae are attracting attention as antihypertensive agents for the prevention of hypertension, which have angiotensin I converting enzyme (ACE) inhibitory action, and are the most actively studied.
Wuppleinori (Porphyra pseudolinearis) is a red algae that is used as a raw material for seaweed products. It is also called Iwa-nori, and is known for its great fragrance. We investigated whether such nori products also contain functional peptides.
-
As with other red algae, Wuppleinori protein is extracted from seaweed powder with water and dialyzed to remove low-molecular-weight components such as salts (preparation method link). Peptides derived from WuppuleInori protein used thermolysin, a protease.
Fig. 1(A) is the result of polyacrylamide electrophoresis (SDS-PAGE). Lane 2 shows protein bands. Lane 3 is the protein hydrolyzed with protease and has no band. Lanes 4 and 5 are fluorescence detection. In the protein lane there is phycobiliprotein fluorescence, while in the hydrolyzate there is fluorescence underneath the migration front.
Figure 1(B) shows the ACE inhibitory activity. Proteins also have ACE inhibitory activity, but it was found that their hydrolysates greatly increased the inhibitory activity.
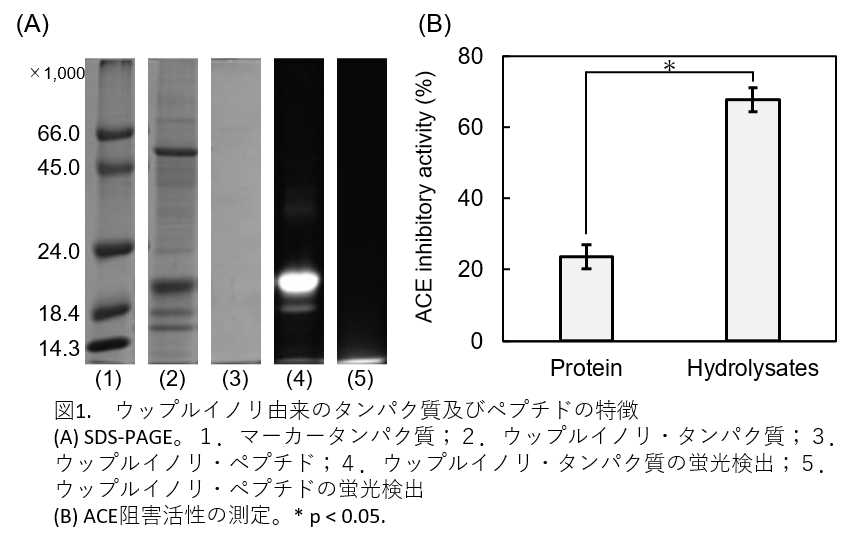
Figure 1. Characteristics of proteins and peptides derived from Wupppleinori
(A) SDS-PAGE. 1. marker protein; 2. Wupppleinori protein; 3. Wupppleinori peptide; 4. fluorescence detection of Wupppleinori protein; 5. fluorescence detection of Wupppleinori peptide
(B) Measurement of ACE inhibitory activity. *p < 0.05.
-
The hydrolyzate contains many peptides. In order to investigate which of these peptides has ACE inhibitory activity, we separated the hydrolyzate by high performance liquid chromatography (HPLC).
Figure 2(A) is the result of separation by HPLC. The numbered peaks in the figure were collected and examined for their ACE inhibitory activity. Figure 2(B) shows the ACE inhibitory activity of each fraction. It can be seen that the strength of activity differs for each fraction. Therefore, we examined what peptides were contained in each fraction using a mass spectrometer.
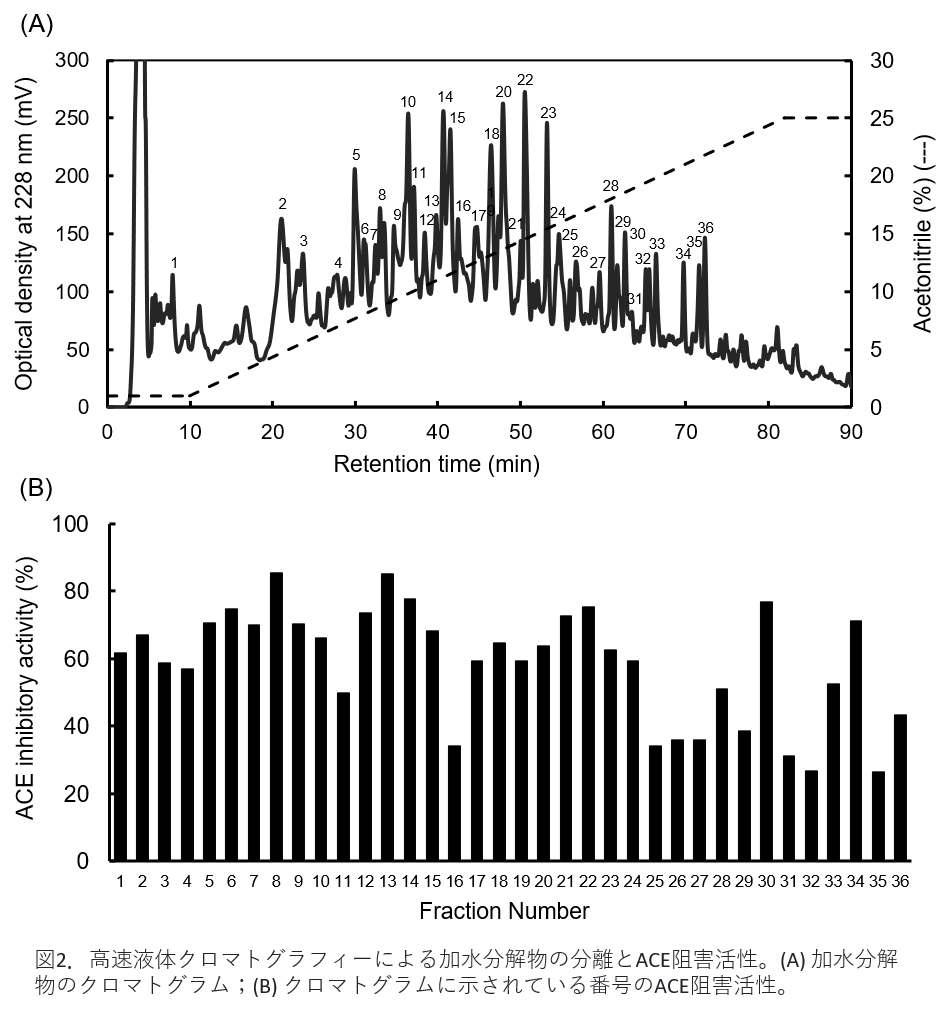
Figure 2. Separation of hydrolyzate and ACE inhibitory activity by high-performance liquid chromatography. (A) chromatograms of hydrolysates; (B) ACE inhibitory activities with numbers indicated in the chromatograms. -
Peptide sequences can be predicted from a mass spectrometer (results can be seen from the linked Table1).
We were able to find 42 peptides. Also, as shown in Figure 1(A), the major protein is phycobiliprotein. To find out if they contain peptide sequences, we examined the protein sequences using next-generation sequencing and polymerase chain reaction (PCR). Table 1 compares the sequences of phycoerythrin (LC599086), phycocyanin (LC599087), and allophycocyanin (LC599088), which are the phycobiliprotein components of upppleinori, with those of other red algae.
As a result, it showed high identity with laver (Pyropia pulchra, Porphyra purpurea, Pyropia yezoensis), and slightly lower identity with dulse (Devaleraea inkyuleei) and centipede laver (Grateloupia asiatica).
Table 1. Identity of the phycobiliproteins of Woodpecker and other red algae
-
Among the 42 peptides, we synthesized 3 peptides (ARY, YLR, LRM) of unknown function derived from phycobiliprotein sequences and examined their ACE inhibitory activity.
As a result, the peptide LRM bound the enzyme most strongly. Its IC50 value is 0.15 μmol. The peptide LRM is a conserved sequence in the alpha chain of phycocyanin. Therefore, we examined how peptide LRM inhibits ACE activity by docking simulation.
Fig. 3(A) shows the peptide bound to ACE. The circles in the figure are zinc ions and are involved in enzyme activity. Peptide LRM shown in purple was shown to suppress the activity by binding to the site that inhibits the binding of zinc ion and the substrate angiotensin I related to blood pressure elevation. Figure 3(B) shows the binding of peptide LRM and ACE in two dimensions.
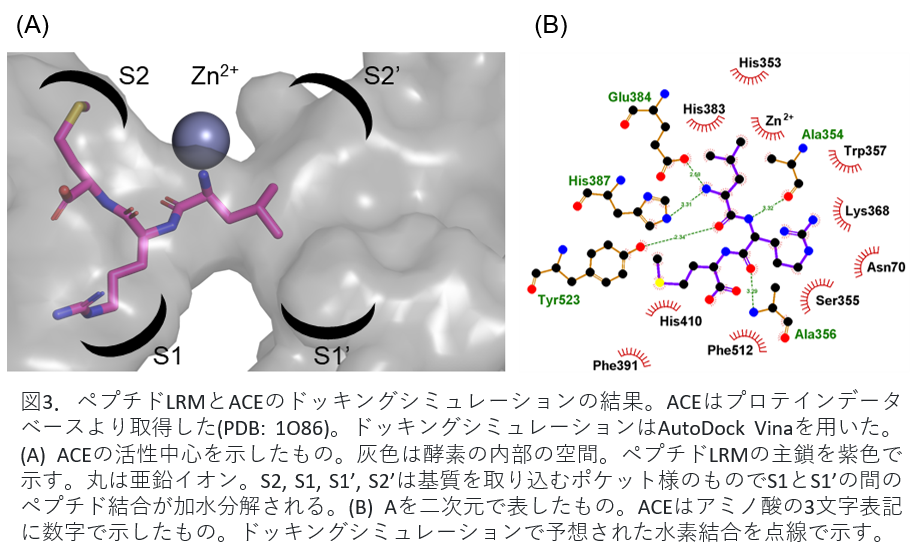
Fig 3. Results of docking simulation of peptide LRM and ACE. ACE was obtained from the protein database (PDB: 1086). AutoDock Vina was used for docking simulation. (A) The active center of ACE is shown. Gray is the internal space of the enzyme. The main chain of peptide LRM is shown in purple. Circles indicate zinc ions; S2, S1, S1', and S2' are pocket-like structures that capture substrates and hydrolyze the peptide bond between S1 and S1'. (B) Two-dimensional representation of A. ACE is a three-letter representation of amino acids with numbers. Hydrogen bonds predicted by docking simulations are indicated by dotted lines.
-
•Search for novel peptides derived from red algae
•Search for functional peptides related to health other than ACE
-




