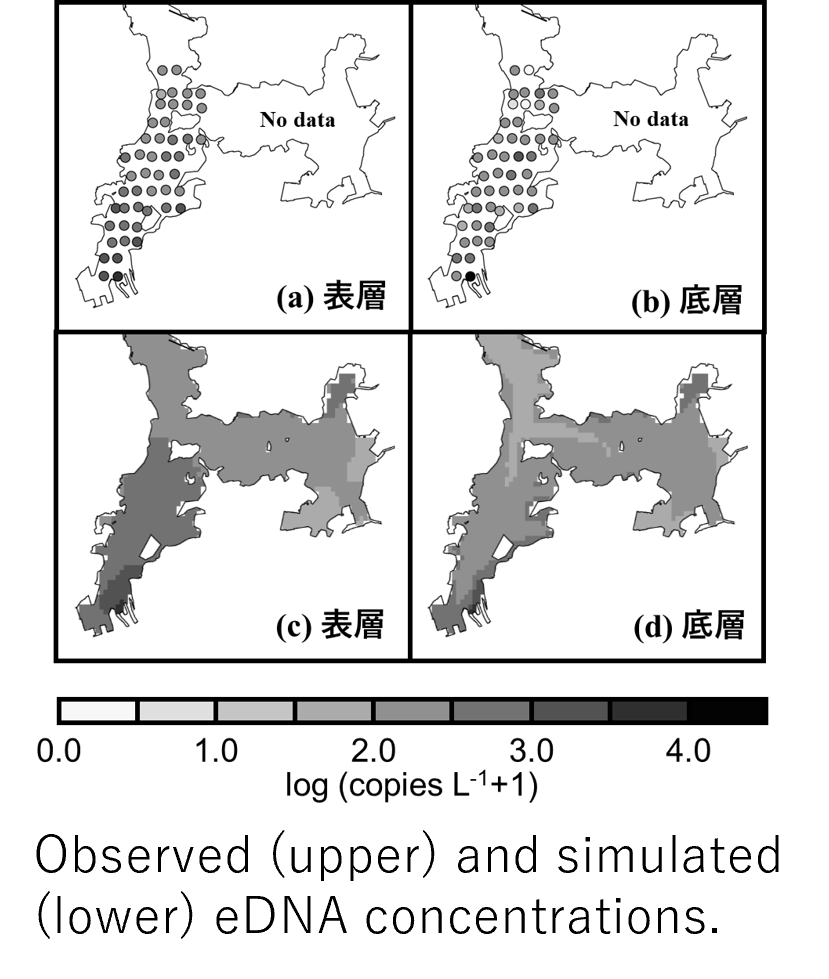
You can analyze the concentration of the eDNA of a specific organism using species-specific primers. It leads to the estimation of biomass without collecting and/or killing organisms. However, the flow is complicated in the sea, and the DNA released from organisms is transported and diffused widely by the flow. In addition, the DNA is gradually decomposed due to the influence of bacteria, ultraviolet rays, and so on. Therefore, we first developed a numerical model that can reproduce the detailed physical process in the water. Applying the model to Maizuru Bay, we simulated the behavior of eDNA released from Japanese horse mackerel. Their distribution was estimated by a fish finder. As a result, we succeeded in reproducing the observed distribution of the eDNA concentration.
Even in the sea, which is not a closed but an open system, the biomass can be estimated from the simulation of the eDNA. We divided the target sea area into fine grids and used the model to simulate how much DNA released from each grid will reach all the grids. Then, we statistically estimated the fish biomass for each grid by calculating the combination to best match the distribution of the actual eDNA concentration. As a result, we obtained similar biomass to that estimated by the fish finder